Want to get Elastic certified? Find out when the next Elasticsearch Engineer training is running! You can start a free cloud trial or try Elastic on your local machine now.
Embedding models output float32 vectors, often too large for efficient processing and practical apps. Elasticsearch supports int8 scalar quantization to reduce vector size while preserving performance. Other methods reduce retrieval quality and are impractical for real world use. In Elasticsearch 8.16 and Lucene, we introduced Better Binary Quantization (BBQ), a new approach developed from insights drawn from a recent technique - dubbed “RaBitQ” - proposed by researchers from Nanyang Technological University, Singapore.
BBQ is a leap forward in quantization for Lucene and Elasticsearch, reducing float32 dimensions to bits, delivering ~95% memory reduction while maintaining high ranking quality. BBQ outperforms traditional approaches like Product Quantization (PQ) in indexing speed (20-30x less quantization time), query speed (2-5x faster queries), with no additional loss in accuracy.
In this blog, we will explore BBQ in Lucene and Elasticsearch, focusing on recall, efficient bitwise operations, and optimized storage for fast, accurate vector search.
Note, there are differences in this implementation than the one proposed by the original RaBitQ authors. Mainly:
- Only a single centroid is used for simple integration with HNSW and faster indexing
- Because we don't randomly rotate the codebook we do not have the property that the estimator is unbiased over multiple invocations of the algorithm
- Rescoring is not dependent on the estimated quantization error
- Rescoring is not completed during graph index search and is instead reserved only after initial estimated vectors are calculated
- Dot product is fully implemented and supported. The original authors focused on Euclidean distance only. While support for dot product was hinted at, it was not fully considered, implemented, nor measured. Additionally, we support max-inner product, where the vector magnitude is important, so simple normalization just won't suffice.
What does the "better" in Better Binary Quantization mean?
In Elasticsearch 8.16 and in Lucene, we have introduced what we call "Better Binary Quantization". Naive binary quantization is exceptionally lossy and achieving adequate recall requires gathering 10x or 100x additional neighbors to rerank. This just doesn't cut it.
In comes Better Binary Quantization! Here are some of the significant differences between Better Binary Quantization and naive binary quantization:
- All vectors are normalized around a centroid. This unlocks some nice properties in quantization.
- Multiple error correction values are stored. Some of these corrections are for the centroid normalization, some are for the quantization.
- Asymmetric quantization. Here, while the vectors themselves are stored as single bit values, queries are only quantized down to int4. This significantly increases search quality at no additional cost in storage.
- Bit-wise operations for fast search. The query vectors are quantized and transformed in such a way that allows for efficient bit-wise operations.
Indexing with Better Binary Quantization (BBQ)
Indexing is simple. Remember, Lucene builds individual read only segments. As vectors come in for a new segment the centroid is incrementally calculated. Then once the segment is flushed, each vector is normalized around the centroid and quantized.
Here is a small example:
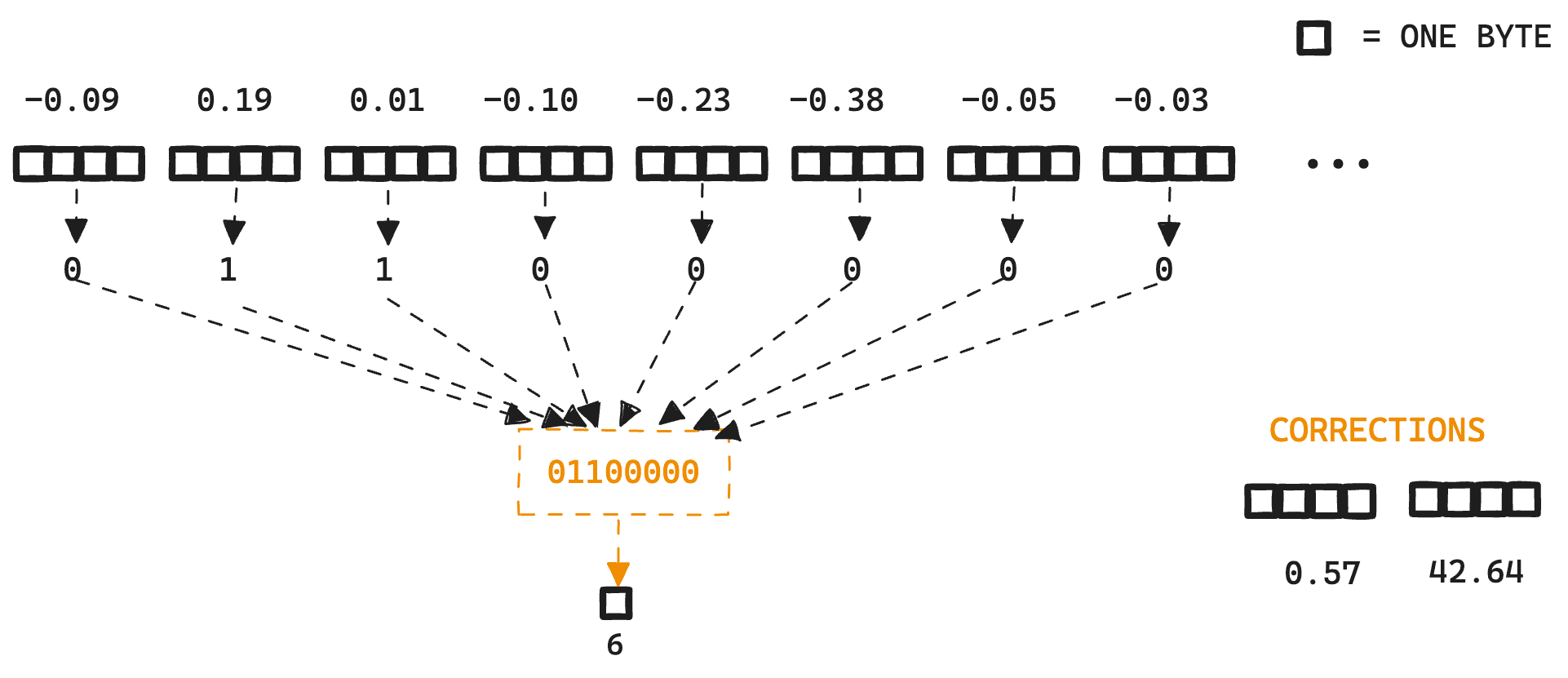
When quantizing down to the bit level, 8 floating point values are transformed into a single 8bit byte.
Then, each of the bits are packed into a byte and stored in the segment along with any error correction values required for the vector similarity chosen.
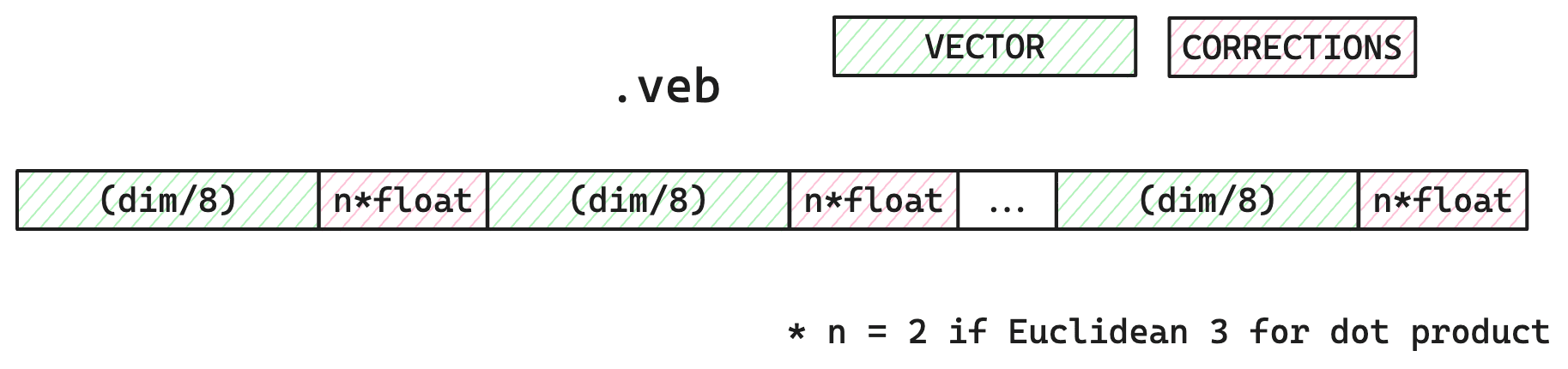
For each vector, bytes stored are dims/8 number of bytes and then any error correction values; 2 floating point values for Euclidean, or 3 for dot product.
A quick side quest to talk about how we handle merging
When segments are merged, we can take advantage of the previously calculated centroids. Simply doing a weighted average of the centroids and then re-quantizing the vectors around the new centroid.
What gets tricky is ensuring HNSW graph quality and allowing the graph to be built with the quantized vectors. What's the point of quantizing if you still need all the memory to build the index?!
In addition to appending vectors to the existing largest HNSW graph, we need to ensure vector scoring can take advantage of asymmetric quantization. HNSW has multiple scoring steps: one for the initial collection of neighbors, and another for ensuring only diverse neighbors are connected. In order to efficiently use asymmetric quantization, we create a temporary file of all vectors quantized as 4bit query vectors.
So, as a vector is added to the graph we first:
- Get the already quantized query vector that is stored in the temporary file.
- Search the graph as normal using the already existing bit vectors.
- Once we have the neighbors, diversity and reverse-link scoring can be done with the previously int4 quantized values.
After the merge is complete, the temporary file is removed leaving only the bit quantized vectors.
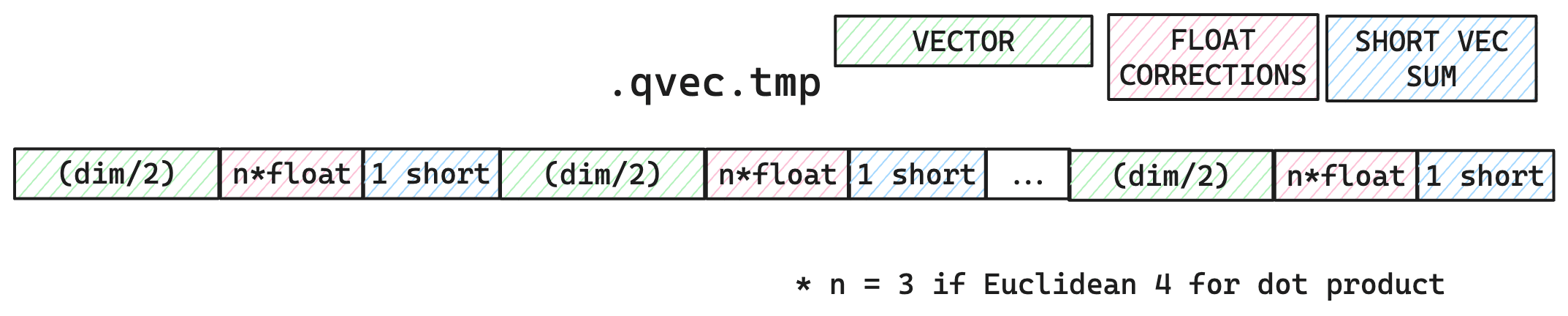
The temporary file stores each query vector as an int4 byte array which takes dims/2 number of bytes, some floating point error correction values (3 for Euclidean, 4 for dot product), and a short value for the sum of the vector dimensions.
Asymmetric quantization, the interesting bits
I have mentioned asymmetric quantization and how we lay out the queries for graph building. But, how are the vectors actually transformed? How does it work?
The "asymmetric" part is straight forward. We quantize the query vectors to a higher fidelity. So, doc values are bit quantized and query vectors are int4 quantized. What gets a bit more interesting is how these quantized vectors are transformed for fast queries.
Taking our example vector from above, we can quantize it to int4 centered around the centroid.
With the quantized vector in hand, this is where the fun begins. So we can translate the vector comparisons to a bitwise dot product, the bits are shifted.
Its probably better to just visualize what is happening:
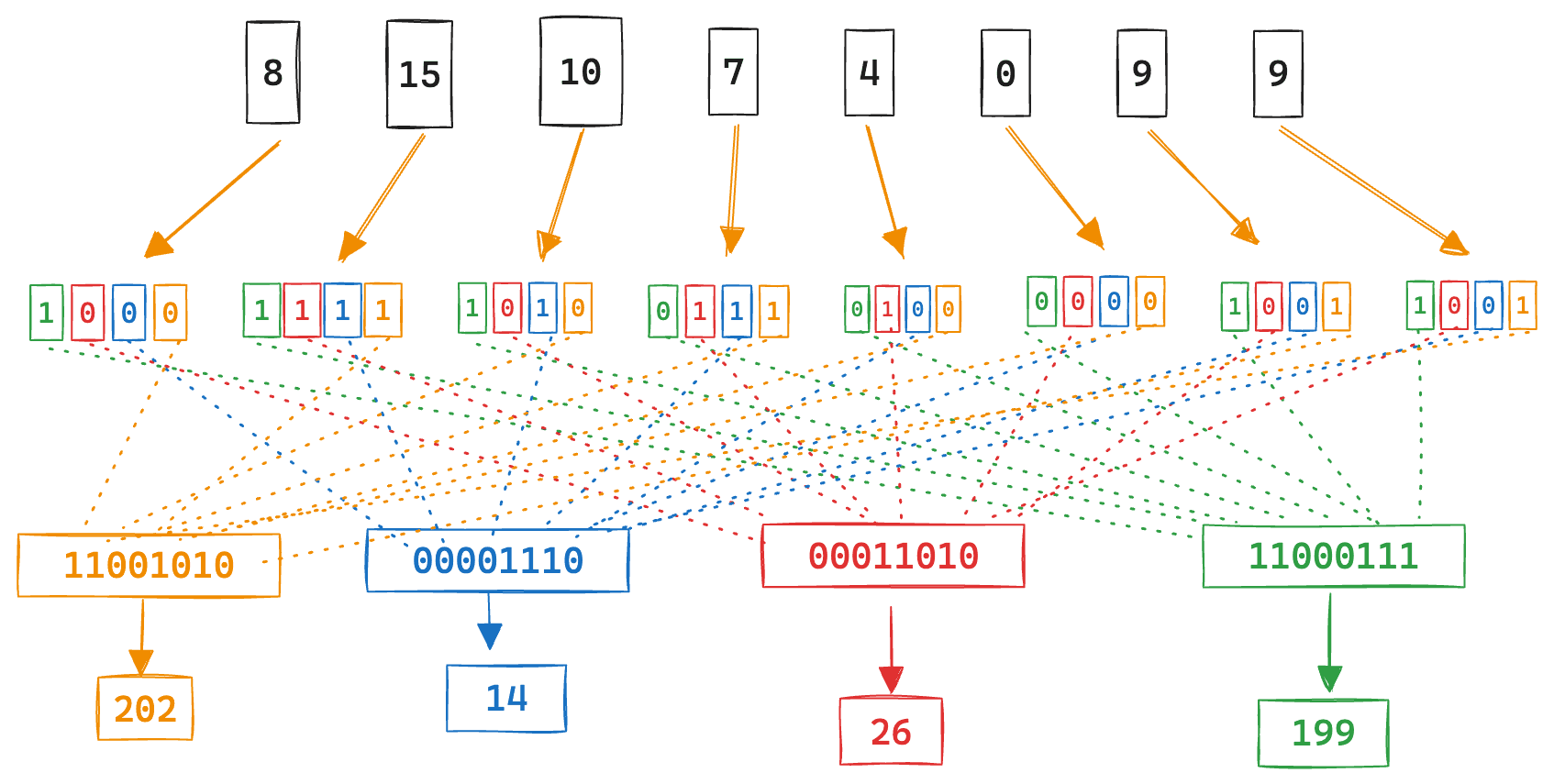
Here, each int4 quantized value has its relative positional bits shifted to a single byte. Note how all the first bits are packed together first, then the second bits, and so on.
But how does this actually translate to dot product? Remember, dot product is the sum of the component products. For the above example, let's write this fully out:
We can see that its simply the summation of the query components where the stored vector bits are 1. And since all numbers are just bits, when expressed using a binary expansion, we can move things around a bit to take advantage of bitwise operations.
The bits that will be flipped after the will be the individual bits of the numbers that contribute to the dot product. In this case 15 and 10.
Now we can count the bits, shift and sum back together. We can see that all the bits that are left over are the positional bits for 15 and 10.
Same answer as summing the dimensions directly.
Here is the example but simplifed java code:
Testing with BBQ: Alright, show me the numbers:
We have done extensive testing with BBQ both in Lucene and Elasticsearch directly. Here are some of the results:
Lucene benchmarking
The benchmarking here is done over three datasets: E5-small, CohereV3, and CohereV2. Here, each element indicates recall@100 with oversampling by [1, 1.5, 2, 3, 4, 5].
E5-small
This is 500k vectors for E5-small built from the quora dataset.
| quantization | Index Time | Force Merge time | Mem Required |
|---|---|---|---|
| bbq | 161.84 | 42.37 | 57.6MB |
| 4 bit | 215.16 | 59.98 | 123.2MB |
| 7 bit | 267.13 | 89.99 | 219.6MB |
| raw | 249.26 | 77.81 | 793.5MB |
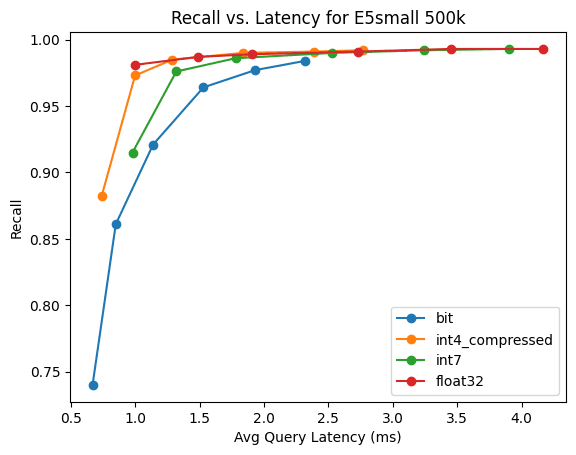
It's sort of mind blowing that we get recall of 74% with only a single bit precision. Since the number of dimensions are fewer, the BBQ distance calculation isn't that much faster than our optimized int4.
CohereV3
This is 1M 1024 dimensional vectors, using the CohereV3 model.
| quantization | Index Time | Force Merge time | Mem Required |
|---|---|---|---|
| bbq | 338.97 | 342.61 | 208MB |
| 4 bit | 398.71 | 480.78 | 578MB |
| 7 bit | 437.63 | 744.12 | 1094MB |
| raw | 408.75 | 798.11 | 4162MB |
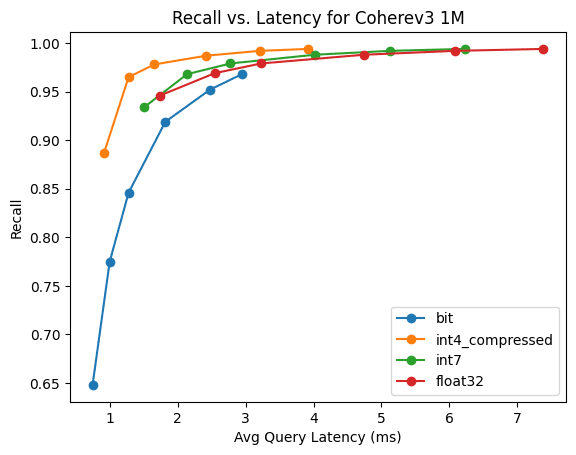
Here, 1bit quantization and HNSW gets above 90% recall with only 3x oversampling.
CohereV2
This is 1M 768 dimensional vectors, using the CohereV2 model and max inner product similarity.
| quantization | Index Time | Force Merge time | Mem Required |
|---|---|---|---|
| bbq | 395.18 | 411.67 | 175.9MB |
| 4 bit | 463.43 | 573.63 | 439.7MB |
| 7 bit | 500.59 | 820.53 | 833.9MB |
| raw | 493.44 | 792.04 | 3132.8MB |
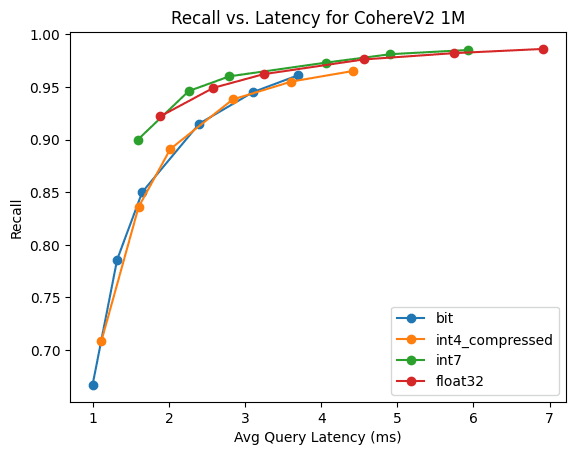
It's really interesting to see how much BBQ and int4 are in lock-step with this benchmark. Its neat that BBQ can get such high recall with inner-product similarity with only 3x oversampling.
Larger scale Elasticsearch benchmarking
As referenced in our larger scale vector search blog we have a rally track for larger scale vector search benchmarking.
This data set has 138M floating point vectors of 1024 dimensions. Without any quantization, this would require around 535 GB of memory with HNSW. With better-binary-quantization, the estimate drops to around 19GB.
For this test, we used a single 64GB node in Elastic cloud with the following track parameters:
Important note, if you want to replicate, it will take significant time to download all the data and requires over 4TB of disk space. The reason for all the additional disk space is that this dataset also contains text fields, and you need diskspace for both the compressed files and their inflated size.
The parameters are as follows:
kis the number of neighbors to search fornum_candidatesis the number of candidates used to explore per shard in HNSWrerankis the number of candidates to rerank, so we will gather that many values per shard, collect the totalreranksize and then rescore the topkvalues with the raw float32 vectors.
For indexing time, it took around 12 hours. And instead of showing all the results, here are three interesting ones:
| k-num_candidates-rerank | Avg Nodes Visited | % Of Best NDGC | Recall | Single Query Latency | Multi-Client QPS |
|---|---|---|---|---|---|
| knn-recall-10-100-50 | 36,079.801 | 90% | 70% | 18ms | 451.596 |
| knn-recall-10-20 | 15,915.211 | 78% | 45% | 9ms | 1,134.649 |
| knn-recall-10-1000-200 | 115,598.117 | 97% | 90% | 42.534ms | 167.806 |
This shows the importance of balancing recall, oversampling, reranking and latency. Obviously, each needs to be tuned for your specific use case, but considering this was impossible before and now we have 138M vectors in a single node, it's pretty cool.
Conclusion
Thank you for taking a bit of time and reading about Better Binary Quantization. Originally being from Alabama and now living in South Carolina, BBQ already held a special place in my life. Now, I have more reason to love BBQ!
We will release this as tech-preview in 8.16, or in serverless right now. To use this, just set your dense_vector.index_type as bbq_hnsw or bbq_flat in Elasticsearch.
Frequently Asked Questions
What is Elastic BBQ?
BBQ (Better Binary Quantization) is a leap forward in quantization for Lucene and Elasticsearch, reducing float32 dimensions to bits, delivering ~95% memory reduction while maintaining high ranking quality.




